CS-RfA-EVBsd (#RDB06090)
Backbone vector plasmid of shRNA expressing lentivirus construct by Gateway(R) cloning with Venus and Bsd marker driven by EF-1 alpha promoter.
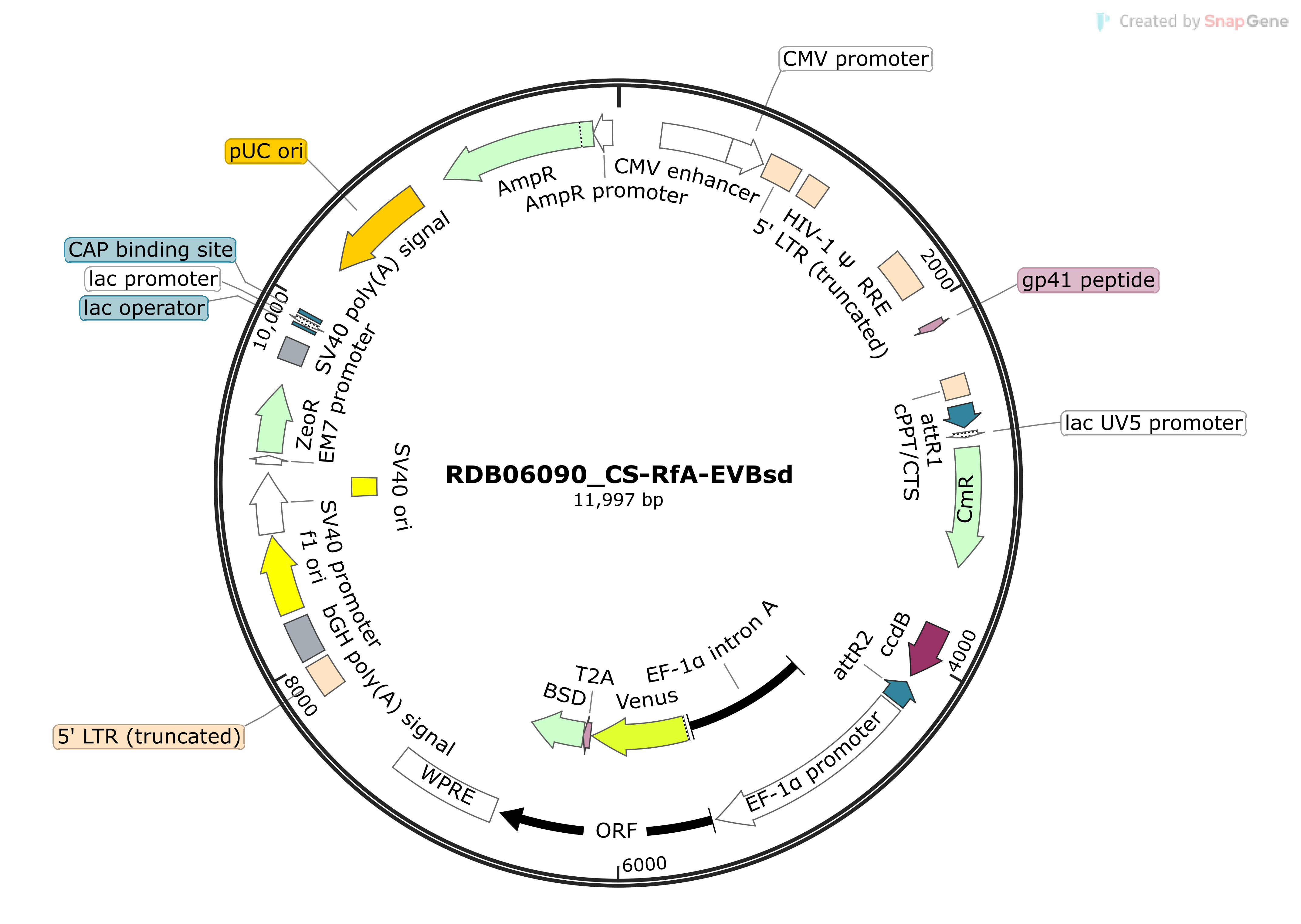 Drawn by SnapGene® software |
|
| Clone info. | Backbone vector plasmid of shRNA expressing lentivirus construct by Gateway(R) cloning with Venus and Bsd marker driven by EF-1 alpha promoter. |
|---|---|
| Comment | Commonly requested with pENTR4-H1 (RDB04395) for generation of shRNA expression vector and with pCMV-VSV-G-RSV-Rev (RDB04393) and pCAG-HIVgp (RDB04394) for packaging. |
| Vector backbone | Plasmid |
| Selectable markers | Ampicillin, Chloramphenicol, ccdB (E. coli), Bsd (lentivirus transfection). Please note that Zeo resistance marker is not included in the lentivirus produced from this vector. |
| Growth conditions | 37C, LB+Amp |
| Growth remarks | Use DB3.1, ccdB Survival or equivalent to amplify this clone. 30 degC is fine. |
| Gene/insert name | Aequorea victoria GFP cDNA |
| Depositor|Developer | Miyoshi, Hiroyuki | |
Distribution information
Please check terms and conditions set forth by the depositor, which are specified in the RIKEN BRC Catalog and/or Web Catalog.
| Ordering forms | Order form [Credit Card Payment MTA, for use for not-for-profit academic purpose [Word Please visit Ordering instruction.[link] |
|---|---|
| Terms and conditions for distribution | The availability of the BIOLOGICAL RESOURCE is limited to a RECIPIENT of a not-for profit institution for a not-for-profit academic purpose. The RECIPIENT agrees to expressly describe the late Dr. Hiroyuki Miyoshi as the Developer. Additional terms and conditions: When publishing the results obtained using the BIOLOGICAL RESOURCE, a citation of literature specified by Dr. Atsushi Miyawaki or an acknowledgement to Dr. Miyawaki is requested. Nagai, T. et al., Nat. Biotechnol. 20: 87-90 (2002). |
| Remarks | Remember that you will be working with samples containing infectious virus. Use Survival2 (Invitrogen) or equivalent to amplify this clone. |
 [open/close]
[open/close]| 必要書類 | 提供依頼書 提供同意書 (MTA, 非営利学術目的用)[Word 手続きの概要は、「レンチウイルスベクターの提供申し込み」をご覧ください。 |
|---|---|
| MTAに書く使用条件 | 本件研究材料は、非営利機関の非営利学術研究に限って提供する。本件研究材料を利用した研究結果等を発表する際は、本件研究材料が故三好浩之博士により開発されたことを明示する。 付加的使用条件: When publishing the results obtained using the BIOLOGICAL RESOURCE, a citation of literature specified by Dr. Atsushi Miyawaki or an acknowledgement to Dr. Miyawaki is requested. Nagai, T. et al., Nat. Biotechnol. 20: 87-90 (2002). |
| 備考 | このリソースはウイルス粒子産生用のため、取扱いに注意が必要です。 このクローンの増殖にはSurvival2 (Invitrogen)あるいは同等の宿主を使用してください。 |
| Catalog # | Resource name | Shipping form | Fee |
|---|---|---|---|
| RDB06090 | CS-RfA-EVBsd | DNA solution |
JPY 9,460 (not-for-profit academic purpose) plus cost of shipping containers, dry ice (if required) and shipping charge |
How to cite this biological resource
Materials & Methods section:
| The CS-RfA-EVBsd was provided by the RIKEN BRC through the National BioResource Project of the MEXT, Japan (cat. RDB06090). |
Reference section:
Further references such as user reports and related articles (go to bottom)
References
Original, user report and related articles
| user_report | Yamagishi, M., Mechanisms of action and resistance in histone methylation-targeted therapy. Nature 627 (8002): 221-228 (2024). PMID 38383791. [PubMed] [Article] [RRC of NBRP] |
|---|---|
| user_report | Araki, H., LSD1 defines the fiber type-selective responsiveness to environmental stress in skeletal muscle. Elife 12: e84618 (2023). PMID 36695573. [PubMed] [Article] [RRC of NBRP] |
| user_report | Truong, T.T.K., Arl4c is involved in tooth germ development through osteoblastic/ameloblastic differentiation. Biochem. Biophys. Res. Commun. 679: 167-174 (2023). PMID 37703759. [PubMed] [Article] [RRC of NBRP] |
| user_report | Hada, M., Highly rigid H3.1/H3.2-H3K9me3 domains set a barrier for cell fate reprogramming in trophoblast stem cells. Genes Dev. 36 (1-2): 84-102 (2022). PMID 34992147. [PubMed] [Article] [RRC of NBRP] |
| user_report | Hasegawa, K., YAP signaling induces PIEZO1 to promote oral squamous cell carcinoma cell proliferation. J. Pathol. 253 (1): 80-93 (2021). PMID 32985688. [PubMed] [Article] [RRC of NBRP] |
| user_report | Nakashima, M., Differentiation of Hodgkin lymphoma cells by reactive oxygen species and regulation by heme oxygenase-1 through HIF-1α. Cancer Sci. 112 (6): 2542-2555 (2021). PMID 33738869. [PubMed] [Article] [RRC of NBRP] |
| user_report | Kamil, M., High filamin-C expression predicts enhanced invasiveness and poor outcome in glioblastoma multiforme. Br. J. Cancer. 120 (8): 819-826 (2019). PMID 30867563. [PubMed] [Article] [RRC of NBRP] |
| user_report | Makino, Y., Single cell RNA-sequencing identified Dec2 as a suppressive factor for spermatogonial differentiation by inhibiting Sohlh1 expression. Sci. Rep. 9 (1): 6063 (2019). PMID 30988352. [PubMed] [Article] [RRC of NBRP] |
| user_report | Yamagishi, M., Targeting Excessive EZH1 and EZH2 Activities for Abnormal Histone Methylation and Transcription Network in Malignant Lymphomas. Cell Rep. 29 (8): 2321-2337.e7 (2019). PMID 31747604. [PubMed] [Article] [RRC of NBRP] |
| user_report | Kimura, H., CKAP4 is a Dickkopf1 receptor and is involved in tumor progression. J. Clin. Invest. 126 (7): 2689-2705 (2016). PMID 27322059. [PubMed] [Article] [RRC of NBRP] |
| user_report | Matsumoto, S., A combination of Wnt and growth factor signaling induces Arl4c expression to form epithelial tubular structures. EMBO J., 33 (7): 702-718 (2014). PMID 24562386. [PubMed] [Article] [RRC of NBRP] |
| user_report | Yamagishi M, Polycomb-Mediated Loss of miR-31 Activates NIK-Dependent NF-κB Pathway in Adult T Cell Leukemia and Other Cancers. Cancer Cell, 21 (1): 121-135 (2012). PMID 22264793. [PubMed] [Article] [RRC of NBRP] |
Resource search
Link
Other resources in our bankExternal Database
Aequorea victoria GFP
Reference sequence
Tips
Featured content
- Lentiviral plasmid list (English text)
- Lentivirus Vector (English text)
- Lentivirus Vector (Japanese text)




